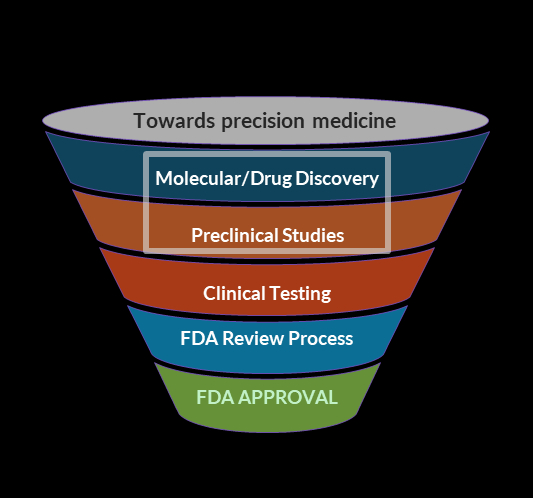
What We Do
Translational research is essential for moving findings from basic life sciences into practical applications. MolVerse increasingly emphasizes on reducing the reliance on animal models in drug development for diseases in humans. Novel in vitro systems, organ-on-a-chip technologies, and advanced AI models serve as alternatives but reducing animal use, and replacing them with more humane approaches in drug discovery is a need since decades. MolVerse accelerates drug discovery pipelines from years to months by providing complete atomic and molecular understanding, rapid compound screening, and precision discovery solutions. We enable better clinical and healthcare decision-making with accurate, efficient and unbiased approach leading to better therapeutic outcomes.
Innovation
Sustainable Innovation through non-invasive methods to understand biomolecular structures, biochemical interactions into biological and disease pathways.
Discovery Solutions
Making structural biology accessible for genomics and proteomics to drug development pipelines and life sciences research.
End-to-End Services
Expedition by reduced data collection and processing time with unbiased machine learning and 3D reconstruction of biological macromolecules or subcellular complexes.
Increased Throughput and Delivery
With our novel reproducible approach, we achieve unparalleled maximum throughput with cost, resources and time efficiency.
Driving Research
Enabling biochemists, healthcare research professionals and hospitals characterize biomarkers, protein complexes and viruses.
Unbiased Structural Solutions
Decipher functional insights through high-resolution structures with precision defying the hypotheses, biases and AI hallucinations.

“When the world is in trouble, chemistry comes to the rescue!”
– Carolyn Bertozzi, Nobel Prize in Chemistry 2022
Unlock The Mysteries of Biomolecular Architectures
- Experience the dynamic molecular architectures and conformations of samples first-hand.
- Shape the future of macromolecular and drug discovery with ease and reproducibility.
- Don’t stop at biochemical functional assays but visualize and take target discovery and knowledge to applications
- Validate biochemical and binding evidence with intricate molecular characterization of the atomic structures.
- Transforming invisible molecular worlds into visible solutions: revealing the hidden language of proteins beyond the surface and folds
- From atoms to answers – revealing nature’s blueprints for better drugs and unlocking therapeutic futures
- Beyond the life’s building blocks, gene and protein sequences are the hidden language of life – we translate their architectural secrets into actionable insights
- Visualization of molecular structures highlighting the intricate dynamics of biomolecular architecture.
Capture Heterogeneity
- Structures from homogeneous and heterogeneous samples.
- Tagged or non-tagged samples from in vitro, in vivo, in situ and native environments.
- Unbiased native biomolecular architectures and subcomplexes for precision understanding of targets and therapeutics.
Our novel technology, tailored solutions and comprehensive services cater to a diverse needs of biochemists, cell biologists, drug developers, life sciences and pharmaceutical researchers to vaccinologists and beyond.
Precision structure determination now accessible, affordable, reproducible and scalable
Meet our team
Team MolVerse brings together seasoned professionals with over a decade of specialized expertise across structural biology, molecular characterization, and ML-driven drug discovery for responsible AI models. Our multidisciplinary team combines deep scientific knowledge with business acumen, ensuring we understand both the technical challenges and commercial realities of drug development. With backgrounds spanning biotechnology, bioinformatics, multi-omics, pharmaceutical sciences, and enterprise management, we deliver solutions that are both scientifically rigorous and commercially viable.